NSF EiR: Upstream and Downstream Analysis of Hoxa1 Gene Expression
During this period (Y1), we analyzed the genomic regions identified by ChIP-on-chip experiments in an attempt to discover novel Hoxa1-binding motifs. We also performed time-series RNA-seq of mouse ES cells treated with retinoic acid for 0, 12, 24, 36, 48, and 60 hours to extend our previous RNA-seq studies and to generate a Hoxa1 Gene Regulatory Network (GRN). By performing DNA-binding assays, we extended our initial studies on the Hoxa1 autoregulatory feedback loop. The results obtained during this period were presented at the annual ASCB meeting in Washington, DC last December. Three undergraduate and one graduate students participated in this project (one undergraduate and one graduate student presented an abstract at the ASCB meeting). Students were trained on different laboratory techniques. They learned to perform cell transfections, EMSA assays, RT-PCR, qPCR, analysis of RNA-seq data, and construction of GRNs.
Ugochi Emelogu in the lab. International graduate student from Nigeria.

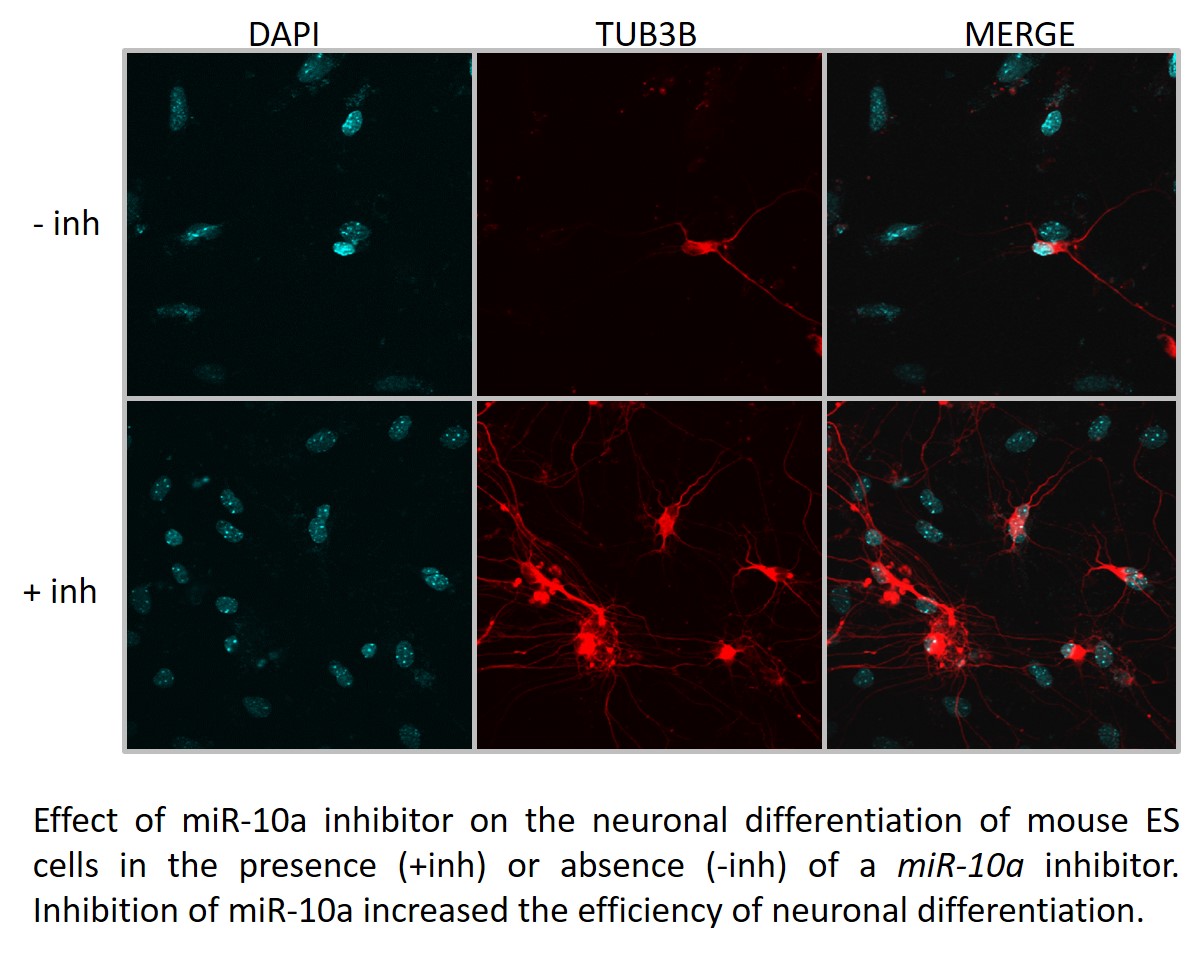
Year 2. We examined the role played by miR-10a, a post-transcriptional repressor of Hoxa1 gene expression, on the neuronal differentiation of mouse ES cells in culture. For this purpose, we performed ES to neuronal differentiation in the presence or absence of a commercial miR-10a inhibitor. transfection of ES cells with the inhibitor, followed by neuronal differentiation using our standard protocol, resulted in about a 4-fold increase in neuronal differentiation efficiency as compared to differentiation in untransfected cells. For future experiments, we will determine whether the different types of neurons are obtained in the presence versus absence of the muR-10a inhibitor.
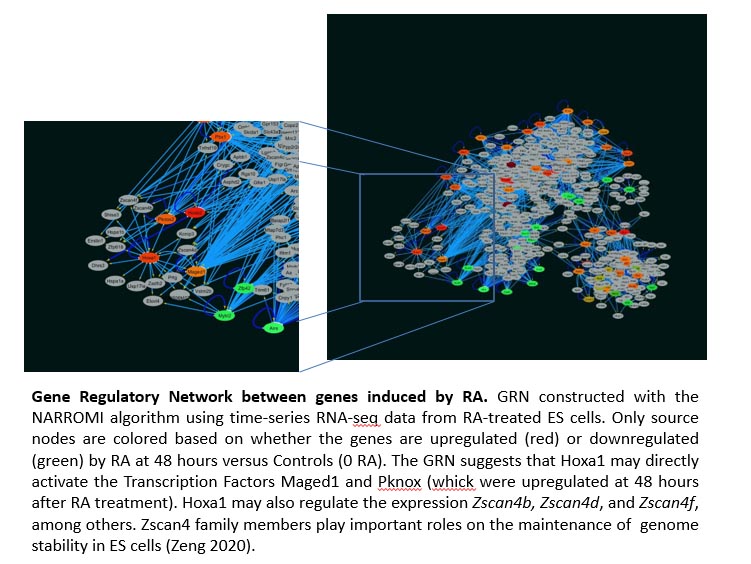
For another aim, we finalized the verification of the time-series RNAseq data used to construct the Hoxa1 Gene Regulatory Network (GRN). The results from the GRN were also verified by qPCR. For this, we examined four genes that directly connect to Hoxa1 according to the GRN. We also examined two genes not connected to Hoxa1, but that connected to Hoxa10 (one of the genes showing dependence on Hoxa1 expression according to the GRN).
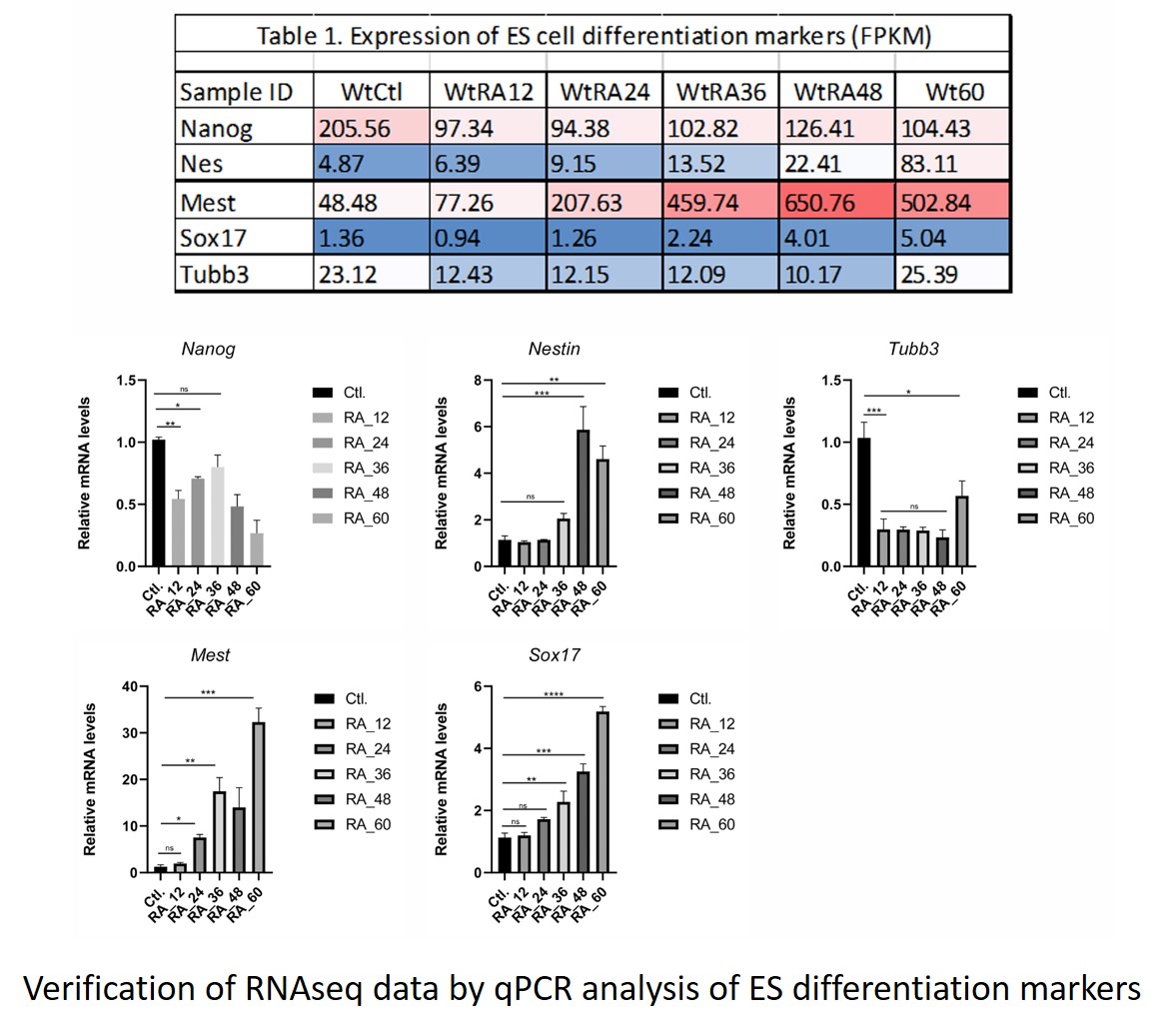
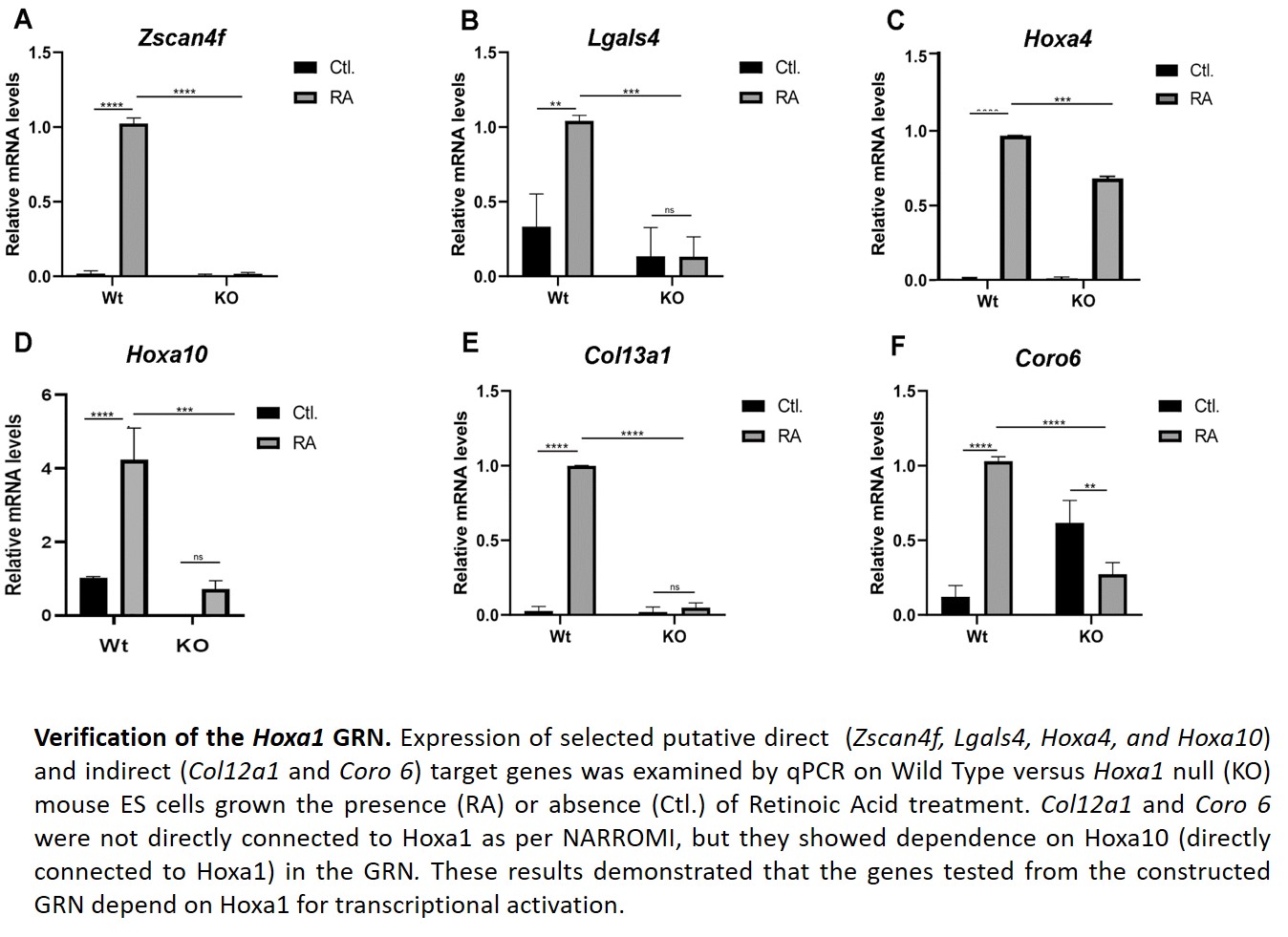
During this period, four undergraduate students and one graduate student participated in the project. They learned bioinformatics techniques and to perform qPCR analyses. One Research Associate joined the laboratory.

Research Associate performing image analyses
Year 3
Major Activities:
During this period, we worked on specific aim 1c seeking to understand the mode of Hoxa1 autoregulation. Specifically, we performed ChIP assays to determine the occupancy status of corepressors in the Hoxa1 locus after treatment of mouse ES cells with Retinoic Acid (RA). We also continued working on specific aim 2 by examining the role of miR-10a on the differentiation of mouse ES cells under 3D culture conditions. With regards to specific aim 3, we performed bulk versus single-cell transcriptome analyses on mouse ES cells grown under two different 3D culture conditions (i.e., Encapsulated vs. Embryoid Bodies). The results obtained from this year’s work were presented by graduate and/or undergraduate students at national and local scientific conferences. One Research Associate, two graduate students, and two undergraduates participated in the project during year 3. One of the undergraduate students graduated, but the rest of the personnel will come back to participate during the coming year. Two additional undergraduate students have expressed interest to join the laboratory and work on this project.
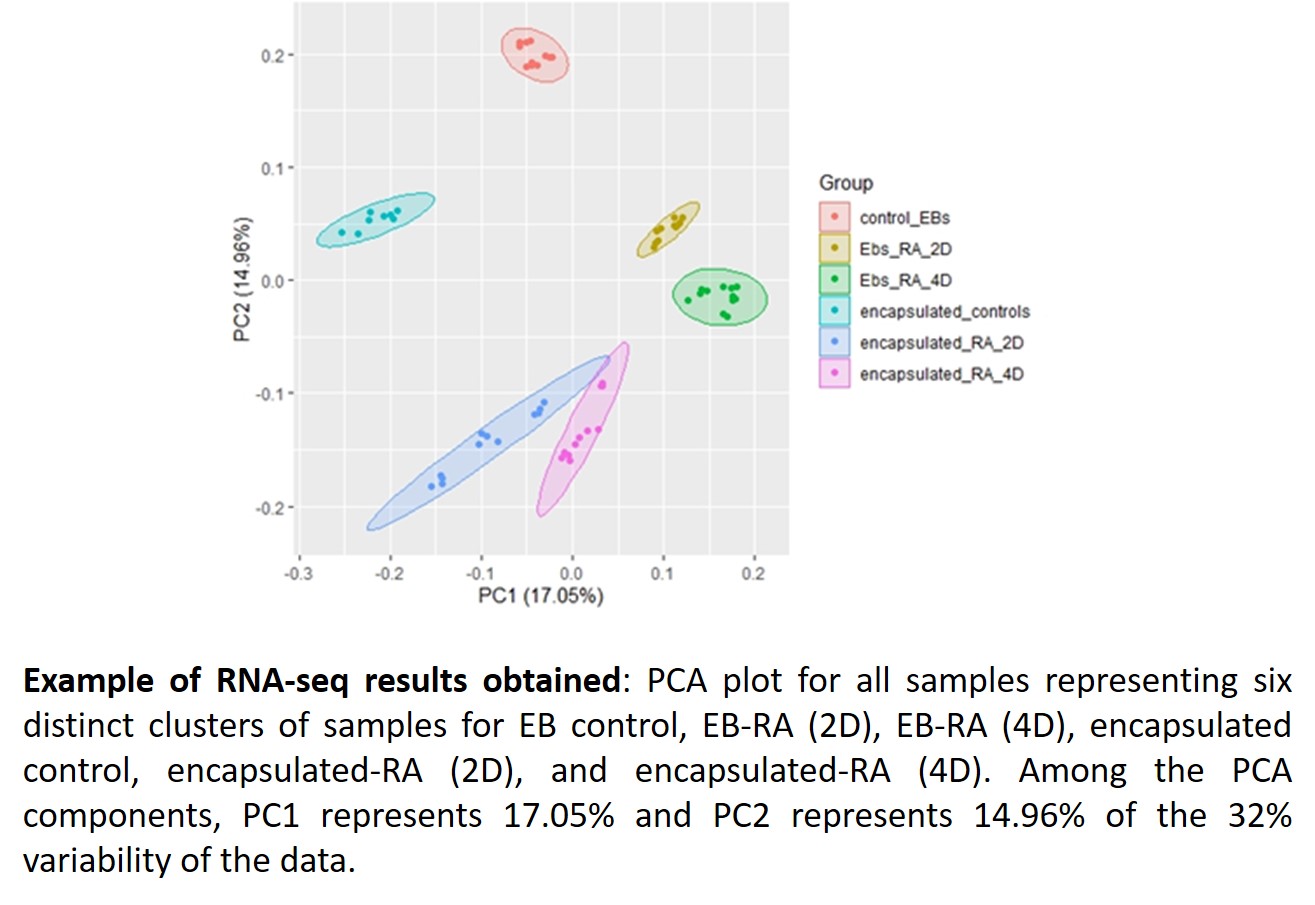
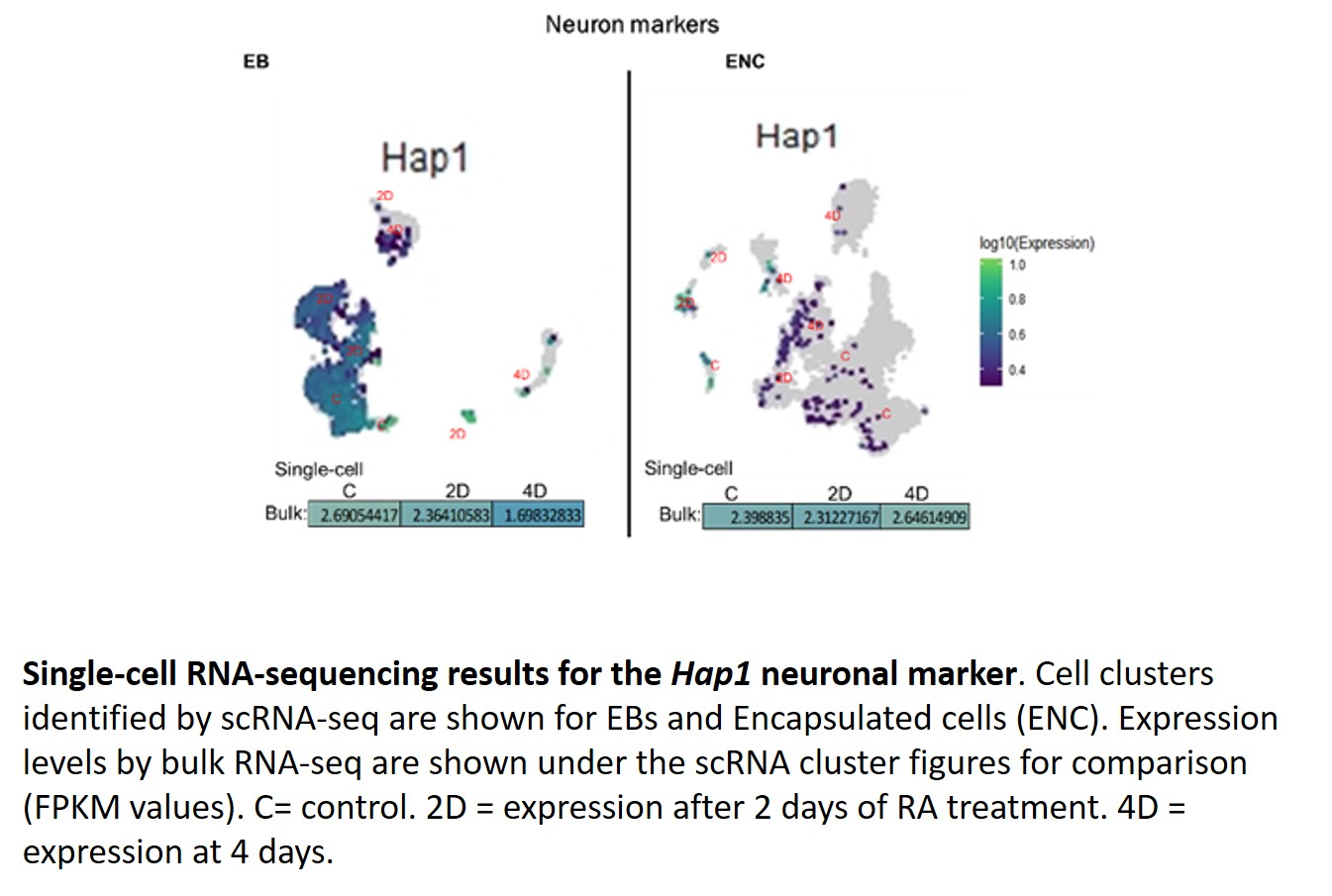
This year, we continued with the characterization of Hoxa1 targets in differentiating ES cells cultured under Encapsulated versus EB 3D culture conditions (Aim 3). As we have previously identified a global set of Hoxa1 target genes, we decided to accomplish this aim by performing time-series bulk versus single-cell RNA sequencing analyses on Encapsulated versus EB mouse ES cells treated with 5 micromolar RA. From our initial analysis of results (see representative figures below), we have found that the following genes were differentially expressed in EBs as compared to Encapsulated cells: Fam46a, Rbp1, Sfrp1, Slc7a3, Ahi1, Hap1, Vegfa, Tubb3, Mafb, and Clu. Particularly, we observed that Hap1, whose gene product plays a role in the delivery of GABAergic receptors to synapses, had decreased expression in EBs as compared to Encapsulated cells, which may be of interest in understanding the molecular mechanisms that direct ES cell differentiation into inhibitory versus excitatory neuronal differentiation.